Human coronavirus 229E
| Human coronavirus 229E (HCoV-229E) | |
|---|---|
 | |
| Videnskabelig klassifikation | |
| Domæne | Riboviria |
| Rige | Orthornavirae |
| Række | Pisuviricota |
| Klasse | Pisoniviricetes |
| Orden | Nidovirales |
| Familie | Coronaviridae |
| Slægt | Alphacoronavirus |
| Underslægt | Duvinacovirus |
| Art | Human coronavirus 229E |
| Hjælp til læsning af taksobokse | |
Human coronavirus 229E ('HCoV-229E') er en art af coronavirus, der inficerer mennesker og flagermus.[1] Det er en indkapslet, enkeltstrenget (single-stranded) RNA-virus med positiv sense, der trænger ind i værtscellen ved at binde til APN-receptoren (aminopeptidase N).[2] Sammen med Human coronavirus OC43 (et medlem af Betacoronavirus-slægten) er det en af de virus, der er ansvarlig for forkølelse.[3][4] HCoV-229E er medlem af slægten Alphacoronavirus og underslægten Duvinacovirus.[5][6]
Referencer
- ^ Lim, Yvonne Xinyi; Ng, Yan Ling; Tam, James P.; Liu, Ding Xiang (2016-07-25). "Human Coronaviruses: A Review of Virus–Host Interactions". Diseases. 4 (3): 26. doi:10.3390/diseases4030026. ISSN 2079-9721. PMC 5456285. PMID 28933406.
See Table 1.
- ^ Fehr AR, Perlman S (2015). Maier HJ, Bickerton E, Britton P (red.). "Coronaviruses: an overview of their replication and pathogenesis". Methods in Molecular Biology. Springer. 1282: 1-23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. PMC 4369385. PMID 25720466.
See Table 1.
− "...Many α-coronaviruses utilize aminopeptidase N (APN) as their receptor, ..." - ^ Susanna K. P. Lau, Paul Lee, Alan K. L. Tsang, Cyril C. Y. Yip,1 Herman Tse, Rodney A. Lee, Lok-Yee So, Yu-Lung Lau, Kwok-Hung Chan, Patrick C. Y. Woo, and Kwok-Yung Yuen. Molecular Epidemiology of Human Coronavirus OC43 Reveals Evolution of Different Genotypes over Time and Recent Emergence of a Novel Genotype due to Natural Recombination. J Virology. 2011 November; 85(21): 11325–11337.
- ^ E. R. Gaunt,1 A. Hardie,2 E. C. J. Claas,3 P. Simmonds,1 and K. E. Templeton. Epidemiology and Clinical Presentations of the Four Human Coronaviruses 229E, HKU1, NL63, and OC43 Detected over 3 Years Using a Novel Multiplex Real-Time PCR Method down-pointing small open triangle. J Clin Microbiol. 2010 August; 48(8): 2940–2947.
- ^ "Virus Taxonomy: 2018 Release". International Committee on Taxonomy of Viruses (ICTV) (engelsk). oktober 2018. Arkiveret fra originalen 20. marts 2020. Hentet 13. januar 2019.
- ^ Woo, Patrick C. Y.; Huang, Yi; Lau, Susanna K. P.; Yuen, Kwok-Yung (2010-08-24). "Coronavirus Genomics and Bioinformatics Analysis". Viruses. 2 (8): 1804-1820. doi:10.3390/v2081803. ISSN 1999-4915. PMC 3185738. PMID 21994708.
Figure 2. Phylogenetic analysis of RNA-dependent RNA polymerases (Pol) of coronaviruses with complete genome sequences available. The tree was constructed by the neighbor-joining method and rooted using Breda virus polyprotein.
Se også
- Virusklassifikation – Definition af termer (virus)
Eksterne henvisninger
 Wikispecies har taksonomi med forbindelse til Human coronavirus 229E
Wikispecies har taksonomi med forbindelse til Human coronavirus 229E- "Human coronavirus 229E, OC43, NL63, HKU1 (RNA) (R-nr. 2058)" hos Ssi.dk, Statens Seruminstitut
- Om Human coronaviruses (bl.a. 229E) hos Virology-online.com, Virology online ("Human coronaviruses were first isolated in the mid 1965 from volunteers at the Common Cold Unit. ..."
- 'Coronaviruses' hos Micro.msb.le.ac.uk, University of Leicester
- Alphacoronavirus hos Viralzone.expasy.org, ViralZone, (SIB Swiss Institute of Bioinformatics)
- "About the Coronaviridae family" hos Viprbrc.org, Virus Pathogen Resource
- 'Taxon identifiers'. Engelsk hjælpeside til 'taksonindentifikatorer'
| Spire Denne artikel om biologi er en spire som bør udbygges. Du er velkommen til at hjælpe Wikipedia ved at udvide den. |
Medier brugt på denne side
Forfatter/Opretter: Emmanuel Boutet, Licens: CC BY-SA 2.5
A black bee (Apis mellifera mellifera.
This illustration, created at the Centers for Disease Control and Prevention (CDC), reveals ultrastructural morphology exhibited by coronaviruses. Note the spikes that adorn the outer surface of the virus, which impart the look of a corona surrounding the virion, when viewed electron microscopically. A novel coronavirus, named Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), was identified as the cause of an outbreak of respiratory illness first detected in Wuhan, China in 2019. The illness caused by this virus has been named coronavirus disease 2019 (COVID-19).
Coronaviruses are a group of viruses that have a halo, or crown-like (corona) appearance when viewed under an electron microscope.
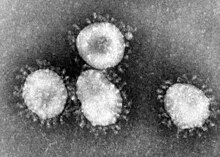
- The coronavirus is now recognized as the etiologic agent of the 2003 SARS outbreak. Additional specimens are being tested to learn more about this coronavirus, and its etiologic link with Severe Acute Respiratory Syndrome. It was previously stated on the CDC website for a coloured version of the image that this depicts the (Avian) Infectious bronchitis virus (IBV), but now states that it depicts Human coronavirus 229E.